 Author
Author  Correspondence author
Correspondence author
Animal Molecular Breeding, 2024, Vol. 14, No. 2
Received: 17 Feb., 2024 Accepted: 31 Mar., 2024 Published: 16 Apr., 2024
This study explores the application of CRISPR-Cas9 gene editing technology to enhance disease resistance in cattle. The CRISPR-Cas9 system has demonstrated significant potential in editing genes to confer disease resistance in livestock. Notable successes include the insertion of the NRAMP1 gene, which has been shown to increase resistance to bovine tuberculosis. The homology-mediated end-joining (HMEJ) method has further improved the efficiency of gene editing in cattle, leading to higher rates of successful gene integration and expression. This study highlights the feasibility and effectiveness of CRISPR-Cas9 in producing disease-resistant livestock. CRISPR-Cas9 gene editing represents a promising tool for enhancing disease resistance in cattle. The technology's ability to precisely modify specific genes associated with disease susceptibility offers a powerful approach to improving livestock health and reducing the economic impact of diseases. By targeting specific genes associated with disease resistance, this study to develop cattle that are more resilient to common and detrimental diseases, thereby improving livestock health and productivity.
1 Introduction
Disease resistance in cattle is a critical trait that significantly impacts livestock productivity, animal welfare, and the economic viability of farming operations. Disease resistance refers to the ability of animals to inhibit the growth of invading pathogens within their bodies, which is influenced by the interaction of the host immune system, host genetics, and the pathogens themselves (Islam et al., 2020; Yuan et al., 2021). The production of disease-resistant cattle can be achieved through molecular breeding techniques that introduce genomic markers responsible for disease resistance or immunocompetence (Islam et al., 2020; Wan et al., 2020).
Despite the potential benefits, enhancing disease resistance in cattle presents several challenges. Traditional breeding methods are often time-consuming and may not always yield the desired results due to the complex nature of genetic traits associated with disease resistance (Wang et al., 2022). Additionally, the occurrence of diseases adversely affects livestock production and animal welfare, impacting both human health and public perception of animal-originated food production. The need for effective disease control approaches has led to the exploration of advanced biotechnological methods, including genome editing, to improve host genetic resistance (Ran et al., 2013; Islam et al., 2020).
CRISPR-Cas9 technology has emerged as a powerful tool for genome editing, enabling precise modifications of the DNA sequence by inserting, deleting, or altering specific genes (Arora and Narula, 2017; Shimatani et al., 2017; Islam et al., 2020). This technology has revolutionized the field of genetic engineering due to its ease of use, high success rate, and cost-effectiveness compared to other genome editing techniques such as zinc-finger nucleases (ZFNs) and transcription activator-like effector nucleases (TALENs) (Hong et al., 2020). The CRISPR-Cas9 system has been successfully applied in various agricultural contexts, including the development of disease-resistant livestock and crops (Borrelli et al., 2018; Wang et al., 2022).
This study comprehensively examines the potential and limitations of CRISPR-Cas9 in livestock disease management by reviewing recent research advancements and relevant case studies, analyzing technological progress in livestock immunogenomics, the principles and applications of CRISPR-Cas9-mediated genome editing, and the commercial prospects of this technology in dairy cattle breeding, aiming to improve animal health and productivity through innovative genetic engineering approaches.
2 CRISPR-Cas9 Technology: Mechanisms and Applications
2.1 Fundamentals of CRISPR-Cas9 gene editing
CRISPR-Cas9, an acronym for Clustered Regularly Interspaced Short Palindromic Repeats and CRISPR-associated protein 9, is a revolutionary genome editing technology that allows for precise, targeted changes to the DNA of living organisms (Figure 1) (Zhang et al., 2021; Tao et al., 2022). Originally discovered as a part of the bacterial immune system, CRISPR-Cas9 has been adapted for use in a wide range of genetic engineering applications. The system works by utilizing a guide RNA (gRNA) to direct the Cas9 enzyme to a specific location in the genome, where it creates a double-strand break. This break can then be repaired by the cell's natural repair mechanisms, allowing for the insertion, deletion, or modification of genes (Liang et al., 2015; Liu et al., 2017; Li et al., 2021).
|
Figure 1 The working mechanism of the CRISPR-Cas system (Adopted from Tao et al., 2022) Image caption: The bacterial defense mechanism of the CRISPR/Cas systems includes three stages: Adaptation stage: acquisition of spacer sequences; Expression stage: Generation of the crRNA and Cas protein; Interference stage: crRNA-guided nucleic acid-targeted cleavage (Adopted from Tao et al., 2022) |
2.2 Applications of CRISPR-Cas9 in animal genetics
CRISPR-Cas9 has been widely adopted in animal genetics for its ability to introduce precise genetic modifications. In livestock, this technology has been used to enhance disease resistance, improve productivity, and introduce desirable traits (Bevacqua et al., 2016). For instance, CRISPR-Cas9 has been employed to insert the NRAMP1 gene in cattle to confer resistance to tuberculosis and to delete the CD163 gene in pigs to make them resistant to porcine reproductive and respiratory syndrome (PRRS) (Islam et al., 2020). Beyond disease resistance, CRISPR-Cas9 is also being explored for its potential to improve other traits such as growth rate, feed efficiency, and meat quality (Barrangou and Doudna, 2016).
2.3 Advantages of CRISPR-Cas9 for disease resistance
The CRISPR-Cas9 system offers several advantages for enhancing disease resistance in livestock. Firstly, it is highly specific and efficient, allowing for precise modifications with minimal off-target effects. This specificity is crucial for ensuring that only the desired genetic changes are made, reducing the risk of unintended consequences (Liang et al., 2015; Liu et al., 2023). Secondly, CRISPR-Cas9 is relatively easy to design and implement compared to other genome editing technologies like TALENs and ZFNs, making it more accessible for widespread use (Borrelli et al., 2018; Eş et al., 2019). Additionally, the decreasing cost of CRISPR-Cas9 technology over time makes it a cost-effective option for genetic improvement programs (Islam et al., 2020; Martínez et al., 2020). Finally, the ability to create transgene-free animals through CRISPR-Cas9-mediated gene editing addresses some of the public and regulatory concerns associated with genetically modified organisms (GMOs) (Langner et al., 2018; Ahmad et al., 2020).
3 Disease Resistance in Cattle: Key Targets for Gene Editing
3.1 Common infectious diseases in cattle
Cattle are susceptible to a variety of infectious diseases that can significantly impact their health and productivity. Some of the most common infectious diseases include Bovine Respiratory Disease (BRD), Bovine Viral Diarrhea (BVD), and Mastitis. These diseases not only affect the well-being of the animals but also lead to substantial economic losses in the dairy and beef industries. Effective management and prevention strategies are crucial to mitigate these impacts.
3.2 Identification of genetic markers associated with disease resistance
The identification of genetic markers associated with disease resistance in cattle is a critical step towards enhancing their resilience through gene editing. Advances in genome sequencing and bioinformatics have enabled the discovery of specific genes and genetic variants that confer resistance to various pathogens. For instance, the eIF4E gene has been identified as a key player in plant virus resistance and could serve as a model for similar studies in cattle (Chandrasekaran et al., 2016). Additionally, the use of high-fidelity CRISPR-Cas9 variants, such as SpCas9-HF1 and eSpCas9 (1.1), has improved the precision of genome editing, reducing off-target effects and increasing the reliability of genetic modifications (Chen et al., 2017).
3.3 CRISPR-Cas9 targets for enhancing resistance to specific diseases
CRISPR-Cas9 technology offers a powerful tool for enhancing disease resistance in cattle by enabling precise modifications of target genes (Mushtaq et al., 2019). For example, the CRISPR-Cas9 system has been successfully used to develop virus-resistant cucumber plants by targeting the eIF4E gene, which could be adapted for similar applications in cattle (Chandrasekaran et al., 2016; Memi et al., 2018). Moreover, the development of new CRISPR-Cas9 variants with enhanced specificity and reduced off-target activity, such as HypaCas9, provides a more accurate approach to gene editing in livestock (Chen et al., 2017). By targeting specific genetic markers associated with disease resistance, CRISPR-Cas9 can be used to create cattle that are more resilient to infectious diseases, thereby improving animal health and productivity.
4 Case Study: CRISPR-Cas9 Mediated Resistance to Bovine Tuberculosis
4.1 Background on bovine tuberculosis
Bovine tuberculosis (bTB) is a chronic infectious disease caused by Mycobacterium bovis, which affects cattle and other animals, including humans. It poses significant economic and public health challenges due to its impact on livestock productivity and its zoonotic potential. Traditional control measures, such as test-and-slaughter policies and vaccination, have limitations, necessitating the exploration of advanced genetic approaches to enhance disease resistance in cattle.
4.2 Identification of resistant genes
The identification of genes associated with resistance to bTB is crucial for developing genetically resistant cattle. Recent advances in immunogenomics have facilitated the identification of candidate genes that play a role in the host immune response to M. bovis infection (Figure 2). One such gene is NRAMP1 (natural resistance-associated macrophage protein 1), which has been implicated in the resistance to various intracellular pathogens, including Mycobacterium species (Islam et al., 2020).
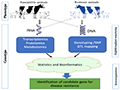 Figure 2 A working pipeline of (in vivo, in vitro, and in silico) immunogenomics for identification of disease resistance candidate gene/marker as prospective targets for genome editing (Adopted from Islam et al., 2020) Image caption: Isolation of single-cell population of a target from both phenotypic groups followed by RNA and DNA extraction separately. The RNA samples could be employed for proteomics and metabolomics profiling. On the other hand, DNA samples could also be subjected to single nucleotide polymorphisms (NSP) sequencing and genotyping, quantitative trait loci (QTL) mapping, and genome-wide association study, and epigenomics study targeting the disease resistance phenotype. Rigorous integrated bioinformatics application on all sets of omics data together enables us to identify the molecular biomarker for the target immunocompetence trait. After functional validation of the identified biomarkers in the independent population, those could be recommended as the targets for CRISPR/Cas9 mediated genome editing technology (Adopted from Islam et al., 2020) |
4.3 Application of CRISPR-Cas9 in developing tuberculosis-resistant cattle
The CRISPR-Cas9 system has emerged as a powerful tool for precise genome editing, enabling the targeted modification of genes associated with disease resistance. In the context of bTB, CRISPR-Cas9 has been employed to insert the NRAMP1 gene into the bovine genome to enhance resistance to M. bovis infection. This approach involves designing single-guide RNAs (sgRNAs) to target specific loci in the bovine genome, followed by the introduction of the Cas9 nuclease to induce double-strand breaks. The NRAMP1 gene is then inserted at the target site through homologous recombination, resulting in the generation of genetically modified cattle with enhanced resistance to bTB (Islam et al., 2020).
4.4 Outcomes and implications of the case study
The application of CRISPR-Cas9 technology to insert the NRAMP1 gene into the bovine genome has shown promising results in developing bTB-resistant cattle. Studies have demonstrated that the edited cattle exhibit a significant reduction in susceptibility to M. bovis infection, as evidenced by lower bacterial loads and improved immune responses compared to non-edited controls (Islam et al., 2020). This genetic approach not only enhances animal health and productivity but also has broader implications for public health by reducing the risk of zoonotic transmission of bTB. Furthermore, the success of this case study underscores the potential of CRISPR-Cas9 technology in advancing livestock disease management and improving food security.
5 Ethical Considerations and Regulatory Framework
5.1 Ethical implications of gene editing in livestock
The application of CRISPR-Cas9 technology in livestock breeding raises several ethical concerns. One primary issue is the welfare of the animals subjected to genetic modifications. While the goal is to enhance disease resistance, the long-term effects on animal health and well-being are not fully understood. Ethical considerations also extend to the potential for unintended genetic consequences, which could affect not only the target animals but also the broader ecosystem if these animals were to interact with wild populations (Islam et al., 2020; Liu et al., 2022).
Moreover, the moral implications of altering the genetic makeup of animals for human benefit must be considered. This includes the debate over whether it is ethically acceptable to manipulate animal genomes to suit human needs, potentially at the expense of the animals' natural characteristics and behaviors (Zhang et al., 2020). The balance between the benefits of disease resistance and the ethical treatment of animals remains a contentious issue that requires ongoing dialogue among scientists, ethicists, and the public.
5.2 Current regulatory landscape for CRISPR-Cas9 in animal breeding
The regulatory framework for CRISPR-Cas9 technology in animal breeding varies significantly across different regions. In some countries, genome-edited animals are subject to the same regulations as genetically modified organisms (GMOs), which involve rigorous safety assessments and approval processes. For instance, the European Union has stringent regulations that classify genome-edited animals as GMOs, requiring comprehensive risk assessments before they can be approved for commercial use (Zhang et al., 2020; Liu et al., 2022).
Other countries, such as the United States, have a more flexible approach. The U.S. Food and Drug Administration (FDA) evaluates genome-edited animals on a case-by-case basis, focusing on the specific genetic changes and their potential impacts rather than the method used to create them. This regulatory landscape is continually evolving as new scientific insights and public opinions emerge, highlighting the need for international harmonization of regulations to facilitate the safe and ethical use of CRISPR-Cas9 in animal breeding (Liu et al., 2022).
5.3 Public perception and acceptance
Public perception and acceptance of CRISPR-Cas9 technology in livestock breeding are crucial for its successful implementation. There is a significant gap between scientific advancements and public understanding, which can lead to skepticism and resistance. Concerns about food safety, environmental impact, and animal welfare are common among the public, and these concerns must be addressed through transparent communication and education (Islam et al., 2020; Zhang et al., 2020).
Engaging with the public to explain the benefits and risks of CRISPR-Cas9 technology, as well as the ethical considerations involved, is essential. Building trust through open dialogue and involving stakeholders in decision-making processes can help mitigate fears and increase acceptance. Additionally, highlighting successful case studies, such as the development of disease-resistant cattle and pigs, can demonstrate the potential positive impacts of this technology on food security and animal health (Islam et al., 2020; Liu et al., 2022).
6 Future Perspectives and Research Directions
6.1 Potential for broader applications of CRISPR-Cas9 in cattle
The CRISPR-Cas9 technology has shown immense potential in various fields, including plant and animal biotechnology. In cattle, the application of CRISPR-Cas9 could extend beyond disease resistance to include traits such as improved milk production, enhanced growth rates, and better feed efficiency. The technology's versatility allows for precise genetic modifications, which can be tailored to address specific needs in cattle breeding programs. For instance, CRISPR-Cas9 has been successfully used to insert the NRAMP1 gene to produce tuberculosis-resistant cattle and to delete the CD163 gene to create pigs resistant to porcine reproductive and respiratory syndrome (PRRS) (Islam et al., 2020). These examples highlight the potential for broader applications of CRISPR-Cas9 in enhancing various economically important traits in cattle.
6.2 Challenges and limitations in current research
Despite the promising potential of CRISPR-Cas9, several challenges and limitations need to be addressed to fully realize its benefits in cattle. One major challenge is the efficiency and precision of the gene-editing process. Off-target effects, where unintended genetic modifications occur, remain a significant concern. Additionally, the delivery of CRISPR-Cas9 components into cattle cells is complex and requires further optimization to ensure high efficiency and minimal off-target activity (Zhu et al., 2020; Li et al., 2021). Another limitation is the regulatory and ethical considerations surrounding the use of gene-editing technologies in livestock. Public perception and acceptance of genetically modified animals can influence the adoption and commercialization of CRISPR-Cas9 applications in cattle breeding (Islam et al., 2020).
6.3 Strategies for enhancing precision and efficiency of gene editing
To overcome the challenges associated with CRISPR-Cas9, several strategies can be employed to enhance the precision and efficiency of gene editing in cattle. One approach is the development of advanced CRISPR-Cas9 variants, such as base editors and prime editors, which allow for more precise genetic modifications with reduced off-target effects (Zhu et al., 2020; Li et al., 2021). Additionally, improving the delivery systems for CRISPR-Cas9 components, such as using viral vectors or nanoparticle-based delivery methods, can enhance the efficiency of gene editing in cattle cells (Langner et al., 2018; Chen et al., 2019). Another strategy is the use of high-throughput screening methods to identify and select the most effective guide RNAs, which can increase the specificity and success rate of CRISPR-Cas9-mediated gene editing (Kershanskaya et al., 2022). By addressing these technical challenges, the precision and efficiency of CRISPR-Cas9 in cattle can be significantly improved, paving the way for its broader application in livestock breeding programs.
7 Concluding Remarks
The application of CRISPR-Cas9 technology in enhancing disease resistance in cattle has shown significant promise. CRISPR-Cas9 has emerged as a powerful tool for genome editing, enabling precise modifications in the DNA sequence to enhance disease resistance in livestock. This technology has been successfully applied to insert or delete specific genes associated with disease resistance, such as the NRAMP1 gene for tuberculosis resistance in cattle. The homology-mediated end-joining (HMEJ)-based method has been demonstrated to increase the efficiency of gene knock-ins compared to traditional homology-directed repair (HDR) methods. This has led to higher success rates in producing gene-edited cattle with predictable expression of functional genes. While the primary focus has been on cattle, CRISPR-Cas9 technology has also been applied to other livestock species, including pigs, goats, and sheep, to enhance disease resistance. This indicates the versatility and broad applicability of the technology in livestock breeding.
CRISPR-Cas9 is poised to play a crucial role in the future of disease resistance in cattle. This technology can make precise genetic modifications. The decreasing cost and increasing technical ease of CRISPR-Cas9 are likely to accelerate its adoption in livestock breeding programs. This will enable the rapid development of disease-resistant cattle, reducing the reliance on traditional breeding methods that are often time-consuming and less precise. The identification of safe harbor loci, such as the ROSA26 locus, ensures that gene insertions do not disrupt other essential genes. This enhances the safety and efficiency of gene editing, making it a viable option for commercial livestock breeding. As the technology continues to advance, it is expected that CRISPR-Cas9 will become a standard tool in livestock breeding. This will not only improve disease resistance but also enhance overall animal welfare and productivity, contributing to sustainable agricultural practices.
The impact of gene editing, particularly through CRISPR-Cas9, on livestock breeding is profound. By enabling precise genetic modifications, this technology offers a revolutionary approach to enhancing disease resistance in cattle and other livestock species. Disease-resistant cattle will experience fewer health issues, leading to better overall welfare and reduced need for antibiotics and other treatments. Healthier livestock translates to higher productivity and lower costs for farmers, contributing to more efficient and profitable farming operations. The ability to breed disease-resistant animals supports sustainable agricultural practices by reducing the environmental impact of livestock farming and ensuring a stable food supply. In conclusion, CRISPR-Cas9 gene editing holds immense potential for transforming livestock breeding. Continued research and development in this field will likely lead to even more innovative applications, further enhancing the health and productivity of cattle and other livestock species.
Acknowledgements
Author would like to express our gratitude to the two anonymous peer reviewers for their critical assessment and constructive suggestions on our manuscript.
Conflict of Interest Disclosure
Author affirms that this research was conducted without any commercial or financial relationships that could be construed as a potential conflict of interest.
Ahmad S., Wei X.J., Sheng Z., Hu P.S., and Tang S., 2020, CRISPR/Cas9 for development of disease resistance in plants: recent progress, limitations and future prospects, Briefings in Functional Genomics, 19(1): 26-39.
https://doi.org/10.1093/bfgp/elz041
PMID: 31915817
Arora L., and Narula A., 2017, Gene editing and crop improvement using CRISPR-Cas9 system, Frontiers in Plant Science, 8: 1932.
https://doi.org/10.3389/fpls.2017.01932
PMID: 29167680 PMCID: PMC5682324
Barrangou R., and Doudna J., 2016, Applications of CRISPR technologies in research and beyond, Nature Biotechnology, 34(9): 933-941.
https://doi.org/10.1038/nbt.3659
PMID: 27606440
Bevacqua R.J., Fernández‐Martín R., Savy V., Canel N.G., Gismondi M.I., Kues W.A., Carlson D.F., Fahrenkrug S.C., Niemann H., Taboga O.A., Ferraris S., and Salamone D.F., 2016, Efficient edition of the bovine PRNP prion gene in somatic cells and IVF embryos using the CRISPR/Cas9 system, Theriogenology, 86(8): 1886-1896.
https://doi.org/10.1016/j.theriogenology.2016.06.010
PMID: 27566851
Borrelli V.M.G., Brambilla V., Rogowsky P., Marocco A., and Lanubile A., 2018, The enhancement of plant disease resistance using CRISPR/Cas9 technology, Frontiers in Plant Science, 9: 1245.
https://doi.org/10.3389/fpls.2018.01245
PMID: 30197654 PMCID: PMC6117396
Chandrasekaran J., Brumin M., Wolf D., Leibman D., Klap C., Pearlsman M., Sherman A., Arazi T., and Gal‐On A., 2016, Development of broad virus resistance in non-transgenic cucumber using CRISPR/Cas9 technology, Molecular Plant Pathology, 17(7): 1140-1153.
https://doi.org/10.1111/mpp.12375
PMID: 26808139 PMCID: PMC6638350
Chen J.S., Dagdas Y.S., Kleinstiver B.P., Welch M.M., Sousa A.A., Harrington L.B., Sternberg S.H., Joung J.K., Yildiz A., and Doudna J.A., 2017, Enhanced proofreading governs CRISPR-Cas9 targeting accuracy, Nature, 550(7676): 407-410.
https://doi.org/10.1038/nature24268
PMID: 28931002 PMCID: PMC5918688
Chen K.L., Wang Y.P., Zhang R.W., Zhang H.W., and Gao C.X., 2019, CRISPR/Cas genome editing and precision plant breeding in agriculture, Annual Review of Plant Biology, 70: 667-697.
https://doi.org/10.1146/annurev-arplant-050718-100049
PMID: 30835493
Eş I., Gavahian M., Martí-Quijal F.J., Lorenzo J.M., Khaneghah A.M., Tsatsanis C., Kampranis S.C., and Barba F.J., 2019, The application of the CRISPR-Cas9 genome editing machinery in food and agricultural science: current status, future perspectives, and associated challenges, Biotechnology Advances, 37(3): 410-421.
https://doi.org/10.1016/j.biotechadv.2019.02.006
Hong Y.H., Meng J., He X.L., Zhang Y.Y., Liu Y.R., Zhang C.W., Qi H.Y., and Luan Y.S., 2020, Editing miR482b and miR482c simultaneously by CRISPR/Cas9 enhanced tomato resistance to Phytophthora infestans, Phytopathology, 111(6): 1008-1016.
https://doi.org/10.1094/PHYTO-08-20-0360-R
PMID: 33258411
Islam M.A., Rony S.A., Rahman M.B., Çınar M.U., Villena J., Uddin M.J., and Kitazawa H., 2020, Improvement of disease resistance in livestock: application of immunogenomics and CRISPR/Cas9 technology, Animals, 10(12): 2236.
https://doi.org/10.3390/ani10122236
PMID: 33260762 PMCID: PMC7761152
Langner T., Kamoun S., and Belhaj K., 2018, CRISPR crops: plant genome editing toward disease resistance, Annual Review of Phytopathology, 56: 479-512.
https://doi.org/10.1146/annurev-phyto-080417-050158
Kershanskaya O.I., Yessenbaeva G.L., Nelidova D.S., Karabekova A.N., and Sadullaeva Z.N., 2022, CRISPR/Cas genome editing perspectives for barley breeding, Physiologia Plantarum, 174(3): e13686.
https://doi.org/10.1111/ppl.13686
PMID: 35451132
Li C., Brant E., Budak H., and Zhang B.H., 2021, CRISPR/Cas: a Nobel Prize award-winning precise genome editing technology for gene therapy and crop improvement, Journal of Zhejiang University-Science B, 22(4): 253-284.
https://doi.org/10.1631/jzus.B2100009
PMID: 33835761 PMCID: PMC8042526
Liang P.P., Xu Y.W., Zhang X.Y., Ding C.H., Huang R., Zhang Z., Lv J., Xie X.X., Chen Y.X., Li Y.J., Sun Y., Bai Y.F., Zhou S.Y., Ma W.B., Zhou C.Q., and Huang J.J., 2015, CRISPR/Cas9-mediated gene editing in human tripronuclear zygotes, Protein & Cell, 6(5): 363-372.
https://doi.org/10.1007/s13238-015-0153-5
PMID: 25894090 PMCID: PMC4417674
Liu H., Ding Y.D., Zhou Y.Q., Jin W.Q., Xie K.B., and Chen L.L., 2017, CRISPR-P 2.0: an improved CRISPR-Cas9 tool for genome editing in plants, Molecular Plant, 10(3): 530-532.
https://doi.org/10.1016/j.molp.2017.01.003
PMID: 28089950
Liu T.F., Ji J., Cheng Y.Y., Zhang S.C., Wang Z.R., Duan K.X., and Wang Y.C., 2023, CRISPR/Cas9-mediated editing of GmTAP1 confers enhanced resistance to Phytophthora sojae in soybean, Journal of Integrative Plant Biology, 65(7): 1609-1612.
https://doi.org/10.1111/jipb.13476
PMID: 36896979
Liu Z.G., Wu T.W., Xiang G.M., Wang H., Wang B.Y., Feng Z., Mu Y.L., and Li K., 2022, Enhancing animal disease resistance, production efficiency, and welfare through precise genome editing, International Journal of Molecular Sciences, 23(13): 7331.
https://doi.org/10.3390/ijms23137331
PMID: 35806334 PMCID: PMC9266401
Martínez M.I.S, Bracuto V., Koseoglou E., Appiano M., Jacobsen E., Visser R.G.F., Wolters A.A., and Bai Y..L, 2020, CRISPR/Cas9-targeted mutagenesis of the tomato susceptibility gene PMR4 for resistance against powdery mildew, BMC Plant Biology, 20(1): 284.
https://doi.org/10.1186/s12870-020-02497-y
PMID: 32560695 PMCID: PMC7304142
Memi F., Ntokou A., and Papangeli I., 2018, CRISPR/Cas9 gene-editing: research technologies, clinical applications and ethical considerations, Seminars in Perinatology, 42(8): 487-500.
https://doi.org/10.1053/j.semperi.2018.09.003
PMID: 30482590
Mushtaq M., Sakina A., Wani S.H., Shikari A.B., Tripathi P., Zaid A., Galla A., Abdelrahman M., Sharma M., Singh A.K., and Salgotra R.K., 2019, Harnessing genome editing techniques to engineer disease resistance in plants, Frontiers in Plant Science, 10: 550.
https://doi.org/10.3389/fpls.2019.00550
PMID: 31134108 PMCID: PMC6514154
Ran F.A., Hsu P.D., Wright J., Agarwala V., Agarwala V., Scott D.A., and Zhang F., 2013, Genome engineering using the CRISPR-Cas9 system, Nature Protocols, 8: 2281-2308.
https://doi.org/10.1038/nprot.2013.143
Shimatani Z., Kashojiya S., Takayama M., Terada R., Arazoe T., Ishii H., Teramura H., Yamamoto T., Komatsu H., Miura K., Ezura H., Nishida K., Ariizumi T., and Kondo A., 2017, Targeted base editing in rice and tomato using a CRISPR-Cas9 cytidine deaminase fusion, Nature Biotechnology, 35(5): 441-443.
https://doi.org/10.1038/nbt.3833
PMID: 28346401
Tao S., Chen H.M., Li N., and Liang W., 2022, The application of the CRISPR-Cas system in antibiotic resistance, Infection and Drug Resistance, 15: 4155-4168.
https://doi.org/10.2147/IDR.S370869
PMID: 35942309 PMCID: PMC9356603
Wan D.Y., Guo Y., Cheng Y., Hu Y., Xiao S.Y., Wang Y.J., and Wen Y.Q., 2020, CRISPR/Cas9-mediated mutagenesis of VvMLO3 results in enhanced resistance to powdery mildew in grapevine (Vitis vinifera), Horticulture Research, 7: 116.
https://doi.org/10.1038/s41438-020-0339-8
PMID: 32821399 PMCID: PMC7395163
Wang S.T., Qu Z.X., Huang Q.Y., Zhang J.F., Lin S., Yang Y.C., Meng F.M., Li J.H., and Zhang K.L., 2022, Application of gene editing technology in resistance breeding of livestock, Life, 12(7): 1070.
https://doi.org/10.3390/life12071070
PMID: 35888158 PMCID: PMC9325061
Yuan M.K., Zhang J.C., Gao Y.P., Yuan Z.K., Zhu Z.L., Wei Y.K., Wu T., Han J., and Zhang Y., 2021, HMEJ-based safe-harbor genome editing enables efficient generation of cattle with increased resistance to tuberculosis, The Journal of Biological Chemistry, 296: 100497.
https://doi.org/10.1016/j.jbc.2021.100497
Zhu H.C., Li C., and Gao C.X., 2020, Applications of CRISPR- Cas in agriculture and plant biotechnology, Nature Reviews Molecular Cell Biology, 21(11): 661-677.
https://doi.org/10.1038/s41580-020-00288-9
Zhang D.B., Hussain A., Manghwar H., Xie K.B., Xie S.S., Zhao S.H., Larkin R.M., Qing P., Jin S.X., and Ding F., 2020, Genome editing with the CRISPR‐Cas system: an art, ethics and global regulatory perspective, Plant Biotechnology Journal, 18(8): 1651-1669.
https://doi.org/10.1111/pbi.13383
PMID: 32271968 PMCID: PMC7336378
Zhang J.F., Khazalwa E.M., Abkallo H.M., Zhou Y., Nie X.W., Ruan J.X., Zhao C.Z., Wang J.R., Xu J., Li X.Y., Zhao S.H., Zuo E.W., Steinaa L., and Xie S.S., 2021, The advancements, challenges, and future implications of the CRISPR/Cas9 system in swine research, Journal of Genetics and Genomics, 48(5): 347-360.
https://doi.org/10.1016/j.jgg.2021.03.015
PMID: 34144928
. FPDF(win)
. FPDF(mac)
. HTML
. Online fPDF
Associated material
. Readers' comments
Other articles by authors
. Xiaofang Lin
Related articles
. CRISPR-Cas9
. Gene editing
. Disease resistance
. Cattle
. Livestock health
Tools
. Post a comment
.png)


